nlmixr2/rxode2 steady state changes
By Matthew Fidler in nlmixr2 rxode2 steady state
April 4, 2024
One of the things that I changed in the last release was steady state.
Once I created the nonmem2rx package, I searched for NONMEM control
streams that we could test the import from, especially those with
attached data. I ran across
nmtests that uses NONMEM to
test against mrgsolve and how it behaves.
During that test I noticed that rxode2 did not follow the convention
of NONMEM in lagged steady state doses. This blog post discusses the
prior and current differences between the steady state of NONMEM and
rxode2/NONMEM.
To reproduce the nmtests in the
current version of rxode2, we will first define the model to compare
with what NONMEM does:
library(rxode2)## rxode2 2.1.2.9000 using 8 threads (see ?getRxThreads)
## no cache: create with `rxCreateCache()`fl <- rxode2({
cl <- 1.1
v <- 20
ka <- 1.5
d/dt(depot) <- -ka*depot
d/dt(central) <- ka*depot - (cl/v)*central
lag(central) <- lagt
f(central) <- bioav
if (mode == 1) rate(central) <- rat2
if (mode == 2) dur(central) <- dur2
cp <- central/(v/1000)
})## using C compiler: ‘gcc (Ubuntu 11.4.0-1ubuntu1~22.04) 11.4.0’
## In file included from /usr/share/R/include/R.h:71,
## from /home/matt/R/x86_64-pc-linux-gnu-library/4.3/rxode2parse/include/rxode2parse.h:33,
## from /home/matt/R/x86_64-pc-linux-gnu-library/4.3/rxode2/include/rxode2.h:9,
## from /home/matt/R/x86_64-pc-linux-gnu-library/4.3/rxode2parse/include/rxode2_model_shared.h:3,
## from rx_54ce7be6c68b8b7855a25655c9d2a337_.c:117:
## /usr/share/R/include/R_ext/Complex.h:80:6: warning: ISO C99 doesn’t support unnamed structs/unions [-Wpedantic]
## 80 | };
## | ^Then we import the test data from nlmixr2data (note this dataset has
been modified from the original
nmtests with a few more tests
cases).
Steady State with Lag time
We will show the first difference we saw in the nmtests:
dfull <- nlmixr2data::nmtest
d <- dfull[dfull$id ==9, ]
print(d[d$evid != 0, ])## id time cp cmt amt evid ii ss addl rate lagt bioav rat2 dur2 mode
## 1057 9 0 1002.7 2 100 1 24 1 3 10 3.16 0.412 10 2 0In this case, there is a lag time for a steady state dose. In the previous version of rxode2 we saw:
library(ggtext)
library(ggplot2)
rxSolve(fl, d, ssAtDoseTime=FALSE) %>%
plot(cp) +
geom_point(data=d, aes(x=time, y=cp), color="red") +
labs(x=NULL,
y=NULL,
title = "<span style = 'color: red;'>NONMEM</span> solve vs <span style='color: black;'>rxode2</span> <span style='font-family: monospace;'>ssAtDoseTime=FALSE</span>",
subtitle="show a difference at dose time until the lag time") +
theme(plot.title = element_markdown()) +
annotate("rect", ymin=c(0, 0), ymax=c(1200, 1200),xmin=c(0, 0), xmax=c(4, 4),
fill="blue", color="blue",
alpha=0.2)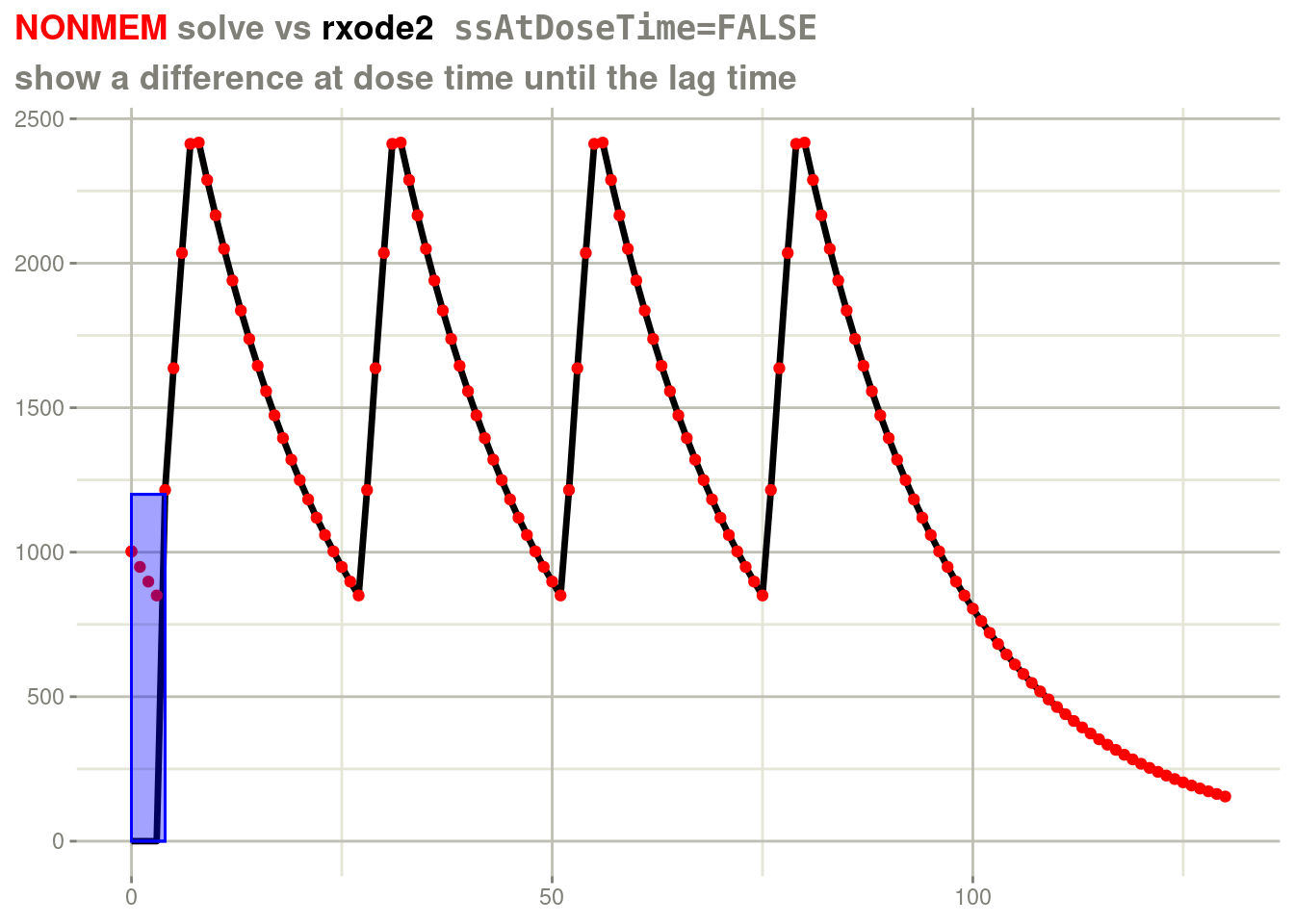
At first I was not concerned with the difference because we do not have to match NONMEM. The purpose of having the steady state in the first place was to simulate steady state. In my opinion, simulating steady state and simulating a few extra doses was the way to make sure you had the correct steady state. (I still think there are situations in NONMEM where this is best practice as I will mention later).
However some in the nlmixr2 team suggested that it should not wait for another cycle, it is more convenient to have the steady state work as expected immediately. I eventually conceded. Hence the new default is to solve back-calculating the steady state of the lag time:
rxSolve(fl, d) %>%
plot(cp) +
geom_point(data=d, aes(x=time, y=cp), color="red") +
labs(x=NULL,
y=NULL,
title = "<span style = 'color: red;'>NONMEM</span> solve vs <span style='color: black;'>rxode2</span> <span style='font-family: monospace;'>ssAtDoseTime=TRUE</span>",
subtitle="show no differences (default as of rxode2 2.1.0)") +
theme(plot.title = element_markdown())
More extreme steady state example
One of the new features of rxode2 is handling steady state where
steady state interval is less than the infusion interval. For
example, from the nmtests
dataset:
dfull <- nlmixr2data::nmtest
d <- dfull[dfull$id ==10, ]
print(d[d$evid != 0, ])## id time cp cmt amt evid ii ss addl rate lagt bioav rat2 dur2 mode
## 1189 10 0 6014.5 2 100 1 12 1 4 2 0 0.812 10 2 0In this case, the infusion lasts well over 12 hours
(100/2*0.812=40.2) even though the steady state dosing is every 12
hours. If this was performed it would mean dosing every 12 hours and
not removing the previous zero order infusion dose. It seems a bit
extreme, but perhaps this is a zero order release tablet that lasts in
the body for a few days. In this case we match NONMEM:
rxSolve(fl, d) %>%
plot(cp) +
geom_point(data=d, aes(x=time, y=cp), color="red") +
labs(x=NULL,
y=NULL,
title = "<span style = 'color: red;'>NONMEM</span> solve vs <span style='color: black;'>rxode2</span> for interdose interval < infusion duration",
subtitle="show no differences (as of rxode2 2.1.0)") +
theme(plot.title = element_markdown())
If you look carefully, there are additional times where each infusion is turned off as the remaining doses wear off.
Another extreme steady state support
In addition to supporting steady state where the inter-dose interval is smaller than the infusion time, we also now support cases where the lag-time is greater than the inter-dose interval. This is an additional test case that is found in our fork of nmtests.
dfull <- nlmixr2data::nmtest
d <- dfull[dfull$id ==409, ]
print(d[d$evid != 0, ])## id time cp cmt amt evid ii ss addl rate lagt bioav rat2 dur2 mode
## 4899 409 0 -18569 2 100 1 24 1 1 10 46 0.412 10 2 0This gives the following:
rxSolve(fl, d) %>%
plot(cp) +
geom_point(data=d, aes(x=time, y=cp), color="red") +
labs(x=NULL,
y=NULL,
title = "<span style = 'color: red;'>NONMEM</span> solve vs <span style='color: black;'>rxode2</span> for lag time > interdose interval",
subtitle="show large at beginning (as of rxode2 2.1.0 and NONMEM 7.4)") +
theme(plot.title = element_markdown())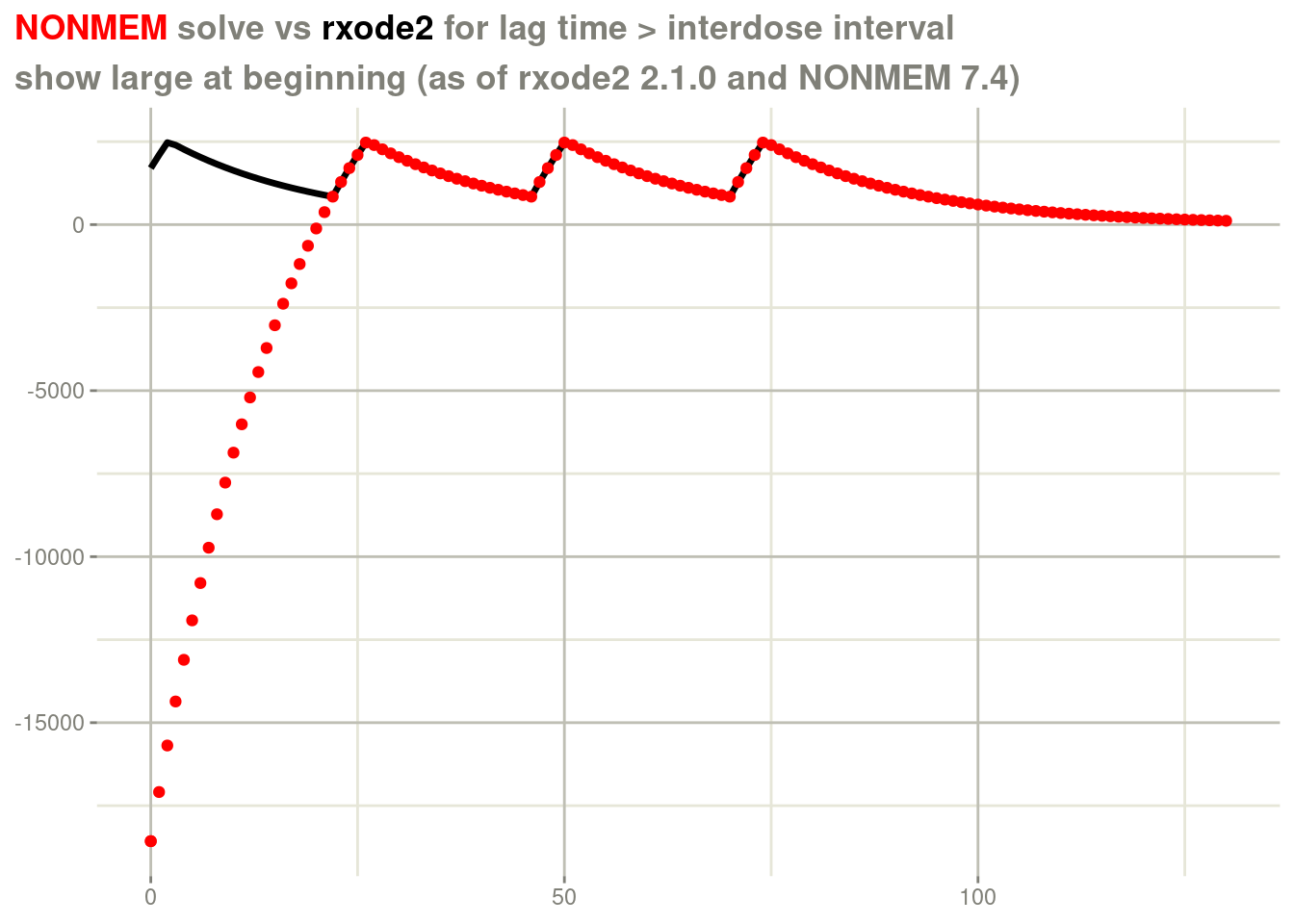
In this case, there are large differences between what NONMEM has and
what rxode2/nlmixr2, especially at the beginning of the profile.
Here NONMEM has negative concentrations. They become positive because
NONMEM changes the steady state doses to non-steady state doses with
the addl flag (as does rxode2). In a few steady state doses after
the initial dose is administered, steady state is achieved in NONMEM
as well. You can see in this plot that the steady state is achieved
immediately in the rxode2 case.
This is one of the reasons why I suggest a few extra doses “just in case”.
I did reach out to Bob Bauer about what I thought was a bug. His reply is as follows:
Rules about steady-state feature are documented as follows:
According to guide VIII, the manual reads, under the section ABSORPTION LAG PARAMETER:
An absorption lag time for a dose is computed by the PK routine using, if needed, information in the dose record. When additional doses are specified on a dose event record, the absorption lag time applies to the dose and to all the additional doses. With a steady-state multiple dose the absorption lag time applies not only to this dose, but also to all the preceding implied doses. With a steady-state dose, the lag time should be less than the interdose interval
In guide VIII, section ADDL DATA ITEM:
For non-steady-state doses, ADDL should be a positive number if and only if the II data item is a positive number. II giv es the time between additional doses. For steady-state infusions, ADDL must be 0. For other steady-state doses, ADDL is optional. If it is a positive number, it continues the pattern of implied doses beyond the steady-state dose. The additional doses of the pattern are non-steady state doses.
And in intro7.pdf:
With NM75 there is a new way of computing SS, the Empirical method, in which there is no SS data item, and a negative value of ADDL requests the computation. This is described separately. (See empirical_SS). (See Guide Introduction_7 “An Empirical Method of Achieving Steady State”)
So my impression is this is not supported by NONMEM (as documented in the manual). The key statement for me is “With a steady-state dose, the lag time should be less than the interdose interval”. However, a run-time error does not raise when this condition occurs (which would be helpful I think).
Note that rxode2 does not currently support the empirical method of
steady state supported in NONMEM 7.5.
Steady state and additional doses
NONMEM supports steady state with additional doses, and as of rxode2
2.1.0, we do too. At first I thought simply duplicating the steady
state records is what was done, but it is not the case.
Lets look at an example where the additional doses take along the steady state flag:
dfull <- nlmixr2data::nmtest
d <- dfull[dfull$id ==25, ]
print(d[d$evid != 0, ])## id time cp cmt amt evid ii ss addl rate lagt bioav rat2 dur2 mode
## 3175 25 0 1891.9 1 100 1 24 1 3 0 0 1 10 2 0
## 3188 25 12 4606.4 1 50 1 24 2 3 0 0 1 10 2 0In this example there is a steady-state 1 followed by a steady state 2 dose. If this solved by duplicated the steady state information you would get:
rxSolve(fl, d, addlDropSs=FALSE) %>%
plot(cp) +
geom_point(data=d, aes(x=time, y=cp), color="red") +
labs(x=NULL,
y=NULL,
title = "<span style = 'color: red;'>NONMEM</span> solve vs <span style='color: black;'>rxode2</span> when <span style='font-family: monospace'>addlDropSs=FALSE</span>",
subtitle="or the steady-state flag is preserved does not match") +
theme(plot.title = element_markdown())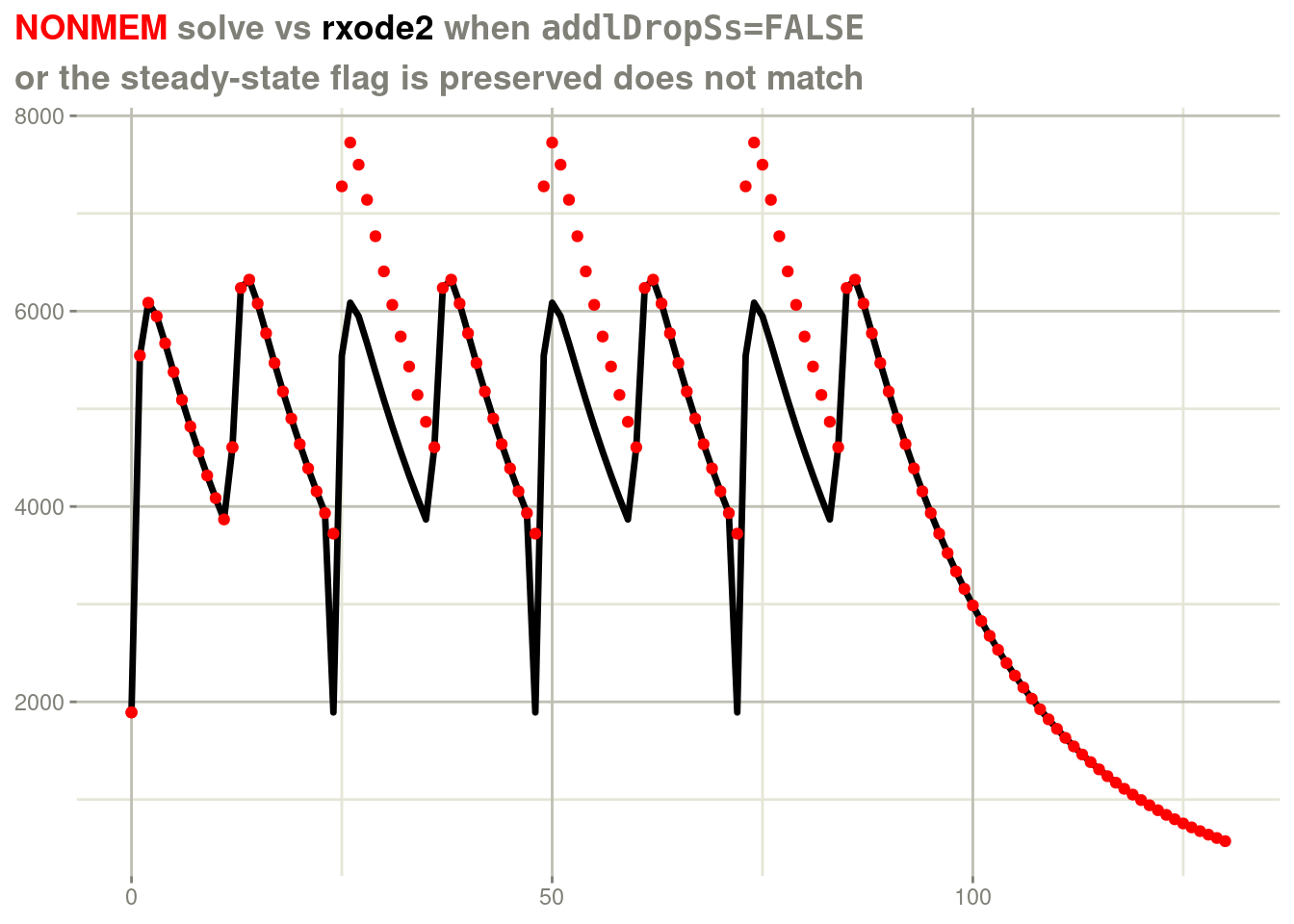
In this case:
The first ODE system is reset and the dose is solved to steady state.
On the second ODE dose, the system is reset and solved to steady state, the old concentration is added to the steady state dose to produce the overall PK curve.
Since the system keeps the steady-state flag with additional doses, this behavior is repeated, which is what you are instructing the system to do, after all.
Note that NONMEM does not keep the steady state flag with additional doses, it simply keeps dosing without the steady state flag. In this case, you would want to keep dosing without the steady-state flag to arrive at the true steady state in about 1 full dosing interval.
This is why the default for rxode2 is to not to keep the steady
state flag either:
rxSolve(fl, d) %>%
plot(cp) +
geom_point(data=d, aes(x=time, y=cp), color="red") +
labs(x=NULL,
y=NULL,
title = "<span style = 'color: red;'>NONMEM</span> solve vs <span style='color: black;'>rxode2</span> when <span style='font-family: monospace'>addlDropSs=TRUE</span>",
subtitle="or the steady-state flag is <span style='color: black'>NOT</span> preserved matches NONMEM") +
theme(plot.title = element_markdown(),
plot.subtitle=element_markdown())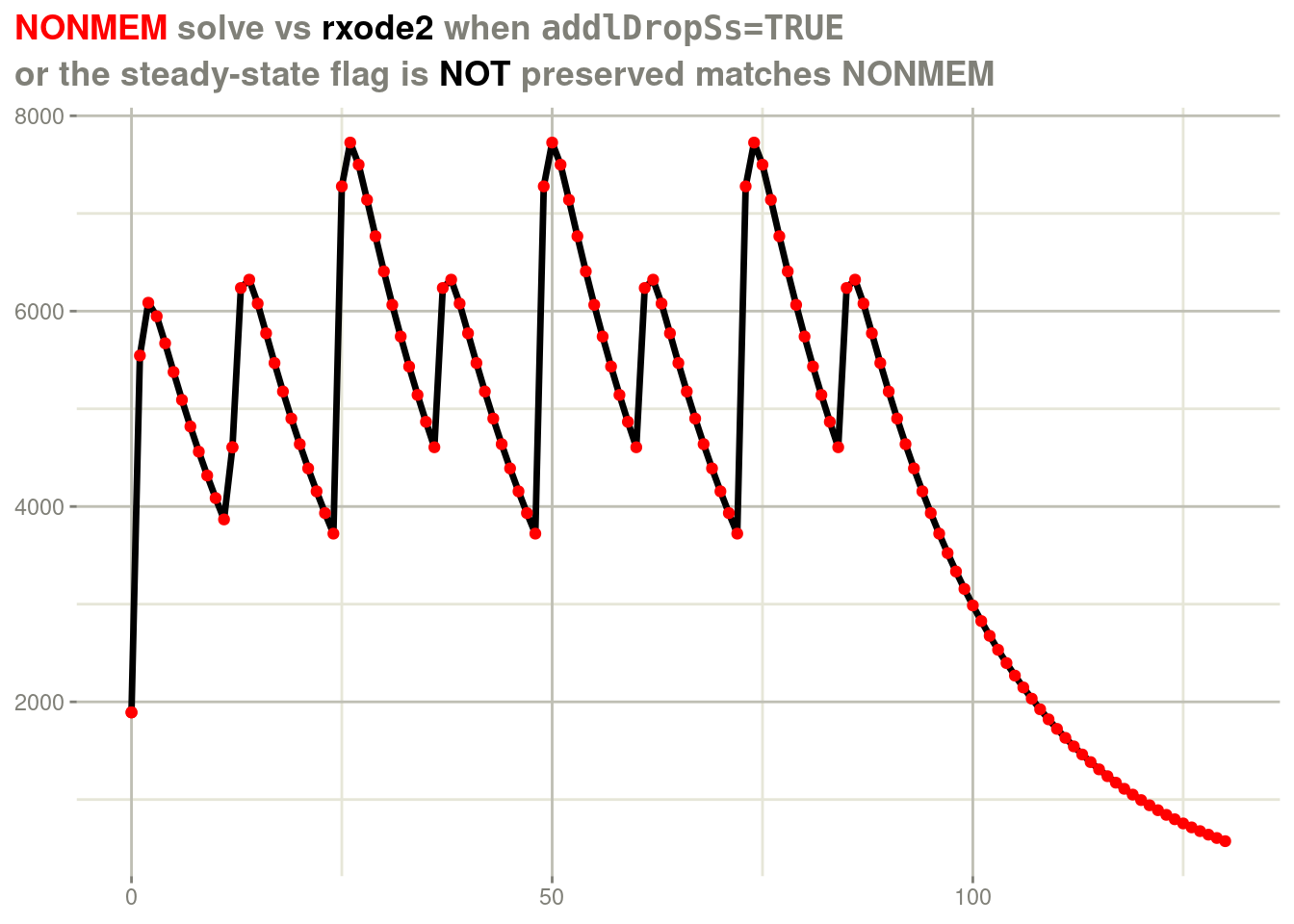
This rolling out of doses without the steady-state flag is also why NONMEM recovers in the prior unsupported steady state scenario.
Other things retained in addl doses in NONMEM vs rxode2
One way that steady state is handled that is different than NONMEM is in covariate definitions:
dfull <- nlmixr2data::nmtest
d <- dfull[dfull$id ==102, ]
print(d[d$evid != 0, ])## id time cp cmt amt evid ii ss addl rate lagt bioav rat2 dur2 mode
## 3440 102 0 0 2 100 1 24 0 3 0 12.13 2.23 10 2 0In this case it is also helpful to see how the covariates are defined:
print(head(d))## id time cp cmt amt evid ii ss addl rate lagt bioav rat2 dur2 mode
## 3440 102 0 0 2 100 1 24 0 3 0 12.13 2.23 10 2 0
## 3441 102 0 0 2 0 0 0 0 0 0 0.00 2.23 10 2 0
## 3442 102 1 0 2 0 0 0 0 0 0 0.00 2.23 10 2 0
## 3443 102 2 0 2 0 0 0 0 0 0 0.00 2.23 10 2 0
## 3444 102 3 0 2 0 0 0 0 0 0 0.00 2.23 10 2 0
## 3445 102 4 0 2 0 0 0 0 0 0 0.00 2.23 10 2 0Here you can see that the covariate lagt starts at 12.13 and then
drops to 0 after the first dose. When comparing NONMEM and rxode2
solves we have the following plot:
rxSolve(fl, d) %>%
plot(cp) +
geom_point(data=d, aes(x=time, y=cp), color="red") +
labs(x=NULL,
y=NULL,
title = "<span style = 'color: red;'>NONMEM</span> solve vs <span style='color: black;'>rxode2</span> when <span style='font-family: monospace'>addlKeepsCov=FALSE</span>",
subtitle="shows a mismatch between NONMEM and rxode2") +
theme(plot.title = element_markdown())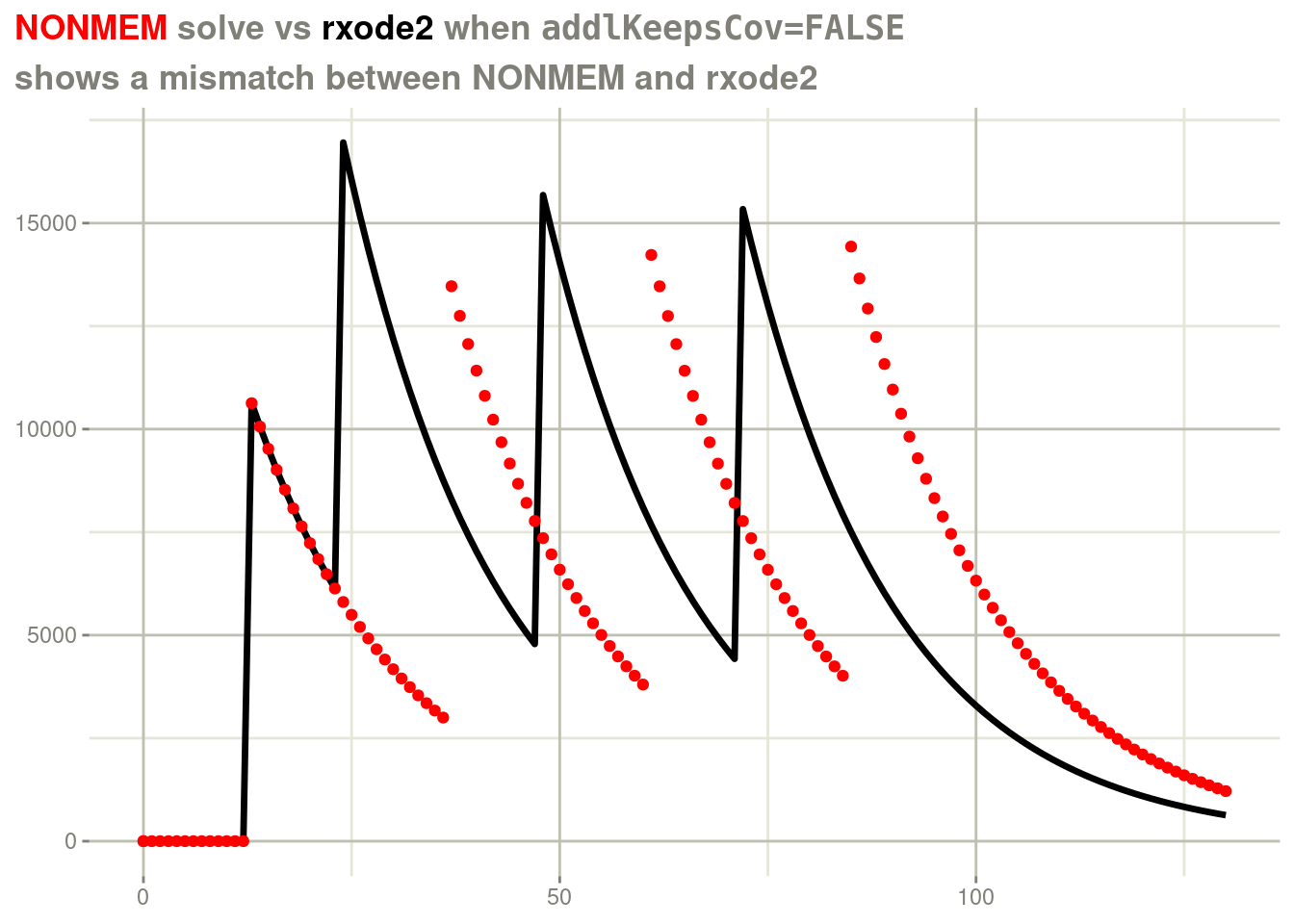
For the rxode2 solve, the covariate lagt is imputed from the
surrounding times instead of carrying forward the value 12.13, which
we at the nlmixr2 team thinks is the right behavior. In contrast,
NONMEM carries the covariate value forward for all of the additional
doses. You can force rxode2 to duplicate the NONMEM way of solving
by addlKeepsCov=TRUE.
rxSolve(fl, d, addlKeepsCov=TRUE) %>%
plot(cp) +
geom_point(data=d, aes(x=time, y=cp), color="red") +
labs(x=NULL,
y=NULL,
title = "<span style = 'color: red;'>NONMEM</span> solve vs <span style='color: black;'>rxode2</span> when <span style='font-family: monospace'>addlKeepsCov=TRUE</span>",
subtitle="shows an exact match between NONMEM and rxode2") +
theme(plot.title = element_markdown())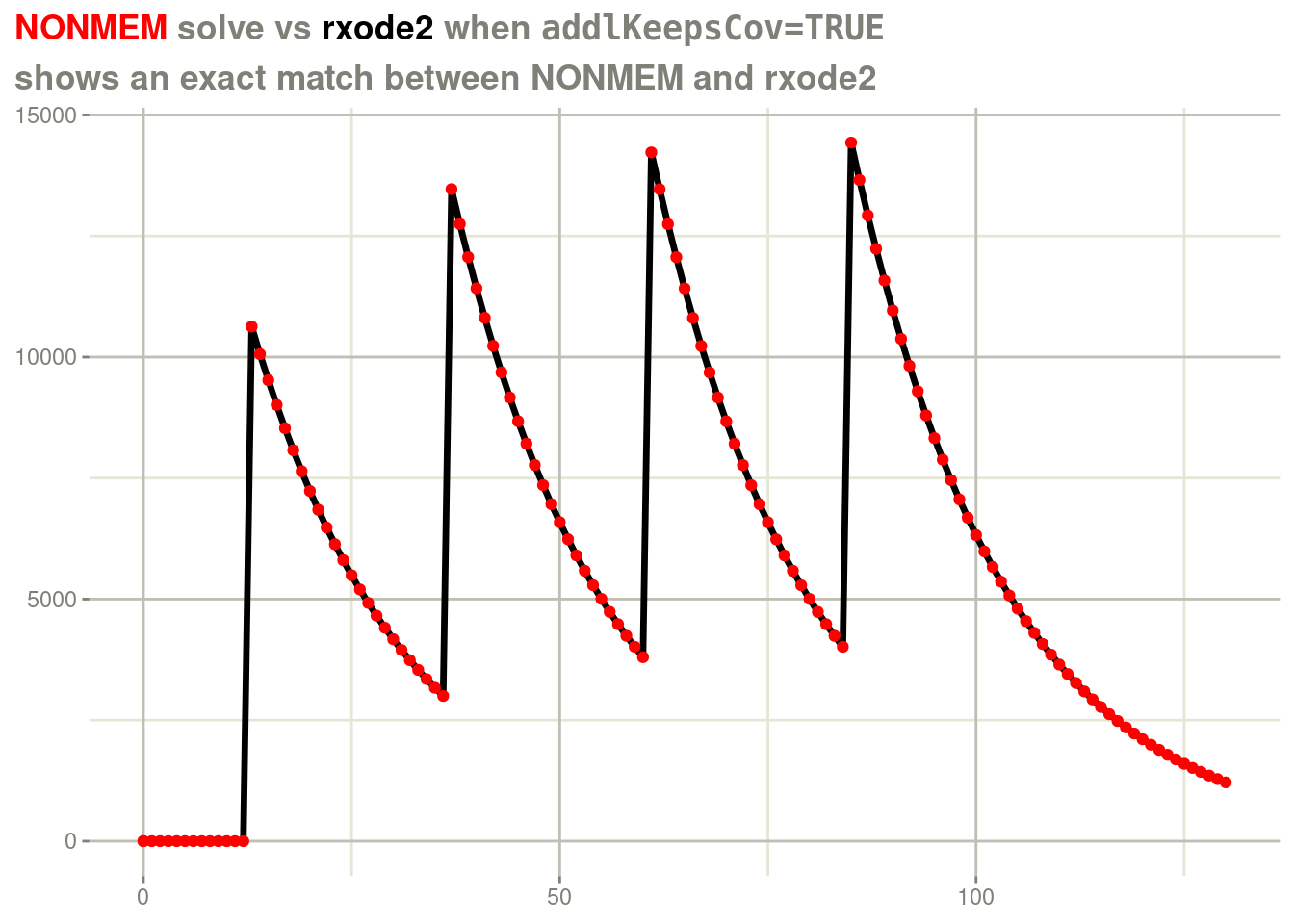
Looking forward
We are working on implementing these same steady state features in the solved systems of rxode2 (and giving the solved systems a general overall by changing the method to get the solved solutions). We decided to push this a bit early because we had to fix some things for CRAN. The old steady state systems still apply for solved linear models (and all the linear model bugs still apply too). We hope to clean these up as soon as we can (though it took us quite a bit of time to implement these steady state features in rxode2 ODE systems).
One of the other things this enables is the ability to do adaptive
dosing inside of rxode2/nlmixr2 which opens up interesting ways of
coding adaptive dosing simulations and maybe dual peak absorption
models.
Conclusion
With this new release of
rxode2,NONMEMandrxode2steady state handling are now closer, but still not the same.We have made some different choices about steady state support (for example the covariate handling, and have not included empirical steady state)
- Posted on:
- April 4, 2024
- Length:
- 11 minute read, 2243 words
- Categories:
- nlmixr2 rxode2 steady state
- Tags:
- steady state
- See Also: