monolix2rx
By Matthew Fidler in rxode2 monolix2rx monolix babelmixr2
October 21, 2024
monolix2rx has been released on CRAN.
I am excited to announce that as part of the nlmixr2 3.0 series of
releases, you now have a way to import a Monolix model into
rxode2/nlmixr2. This is similar to how you can import the output
of nonmem into a rxode2 model with nonmem2rx. I am also in the
process of releasing a bug-fix revision now.
Key notes in importing models to rxode2 and nlmixr2
There are a few things to note in any conversion from monolix to
rxode2 and nlmixr2:
The ODE solvers are different (there is no
lsodasolver inmonolix). This means there are more expected differences in solutions when comparing to importingADVAN13fromNONMEM.rxode2/nlmixr2event tables and event handling are more closely aligned toNONMEMevent tables and event handling. The Monolix data-set definition is different and usesADM, which means when you observe differences between the imported model and the solved model, this can be due data conversion or model conversion. You may want to simulate yourself to see if you can reproduce other known items in the model to validate the model translation is correct.Models in Monolix may not import correctly if they are using the Monolix model library without having
Monolixsetup to read them (details here).When using/sharing/converting Monolix models, you will need to include the model ODE model file, the data (for QCing, optional), the mlxtran file, and the model fit output directory (reading parameter estimates, and output, also optional but can be quite helpful).
Notes about the new matrix specification in lotri
As promised in the last release, I am quickly commenting on why we decided to split the lines to allow the following syntax for matrices:
a ~ 0.2
b ~ c(0.01, 0.2)As noted in the imported matrix with the covariance of the population
parameters, these matrices can be quite large. Because of their size,
without splitting them in an easier to read format it is difficult to
tell the covariance between a and b is 0.01. When the matrices
get large enough, not only do they use this format but they also call
out what parameter is part of the covariance matrix
In the theopylline example, we have one line of the covariance matrix listed as:
a ~ c(omega_ka = -0.0001227, omega_V = -6.5914e-05, omega_Cl = -0.00041194,
a = 0.015333)This covariance matrix now clearly shows that the covariance between
omega_V and a is very small (-6.59e-5). To me, the legibility is
worth the extra method of inputting covariance matrices in lotri.
What this means
With the conversion of a nlmixr2 fit to NONMEM and a nlmixr2 fit
to Monolix, this means that using nlmixr2 you can convert between
NONMEM, nlmixr2 and Monolix making the nlmixr2 a format and
tool you can use to convert models and streamline processes of
pharmacometric modeling.
This is what we are talking about in our free tutorial at ACoP 2024
“Using Past Models to Bridge to Open Models and Open Science using
nlmixr2”: Thursday, November 14, 2024 8:00 AM - 12:00 PM MST Location:
Sonoran Sky Ballroom 1 - 4. Bring your laptops with babelmixr2
setup and you can follow along.
Here we will talk about the conversion process, and some things you
can do with the nlmixr2 model format including optimal design in
PopED (which we have a poster about at ACoP), and individualizing
dosing using posologyr.
Other notes
Note that other things in the nlmixr2 ecosystem have been updated
including dparser (so it will work on older R versions) and
nlmixr2est (adding a new mu-referencing mu-4 that will work with
monolix2rx and other similar models better).
This more frequent release does not have the same impact of making the
whole nlmixr2 ecosystem unstable with the 3.0 release. This means
we will update more frequently since the impact on CRAN is smaller.
Worked example
In this example, we will:
Import a
Monolixmodel torxode2Check the validation of that import simply with
plot(rx)Further convert the
rxode2model to anlmixr2fit object.
library(monolix2rx)
# First load in the model; in this case the theo model
# This is modified from the Monolix demos by saving the model
# file as a text file (hence you can access without model library).
# Additionally some of the file paths were shortened so they could
# be included with monolix2rx
pkgTheo <- system.file("theo", package="monolix2rx")
mlxtranFile <- file.path(pkgTheo, "theophylline_project.mlxtran")
# Note in your own project, you would simply use the file path, you do
# not have to use `system.file()`. This is only because you are loading
# the monlix model from the `monolix2rx` package.
rx <- monolix2rx(mlxtranFile)## ℹ integrated model file 'oral1_1cpt_kaVCl.txt' into mlxtran object## ℹ updating model values to final parameter estimates## ℹ done## ℹ reading run info (# obs, doses, Monolix Version, etc) from summary.txt## ℹ done## ℹ reading covariance from FisherInformation/covarianceEstimatesLin.txt## ℹ done## Warning in .dataRenameFromMlxtran(data, .mlxtran): NAs introduced by coercion## ℹ imported monolix and translated to rxode2 compatible data ($monolixData)## ℹ imported monolix ETAS (_SAEM) imported to rxode2 compatible data ($etaData)## ℹ imported monolix pred/ipred data to compare ($predIpredData)## using C compiler: ‘gcc (Ubuntu 11.4.0-1ubuntu1~22.04) 11.4.0’## ℹ solving ipred problem## ℹ done## ℹ solving pred problem## ℹ donerx## ── rxode2-based free-form 2-cmt ODE model ──────────────────────────────────────────────────────
## ── Initalization: ──
## Fixed Effects ($theta):
## ka_pop V_pop Cl_pop a b
## 0.42699448 -0.78635157 -3.21457598 0.43327956 0.05425953
##
## Omega ($omega):
## omega_ka omega_V omega_Cl
## omega_ka 0.4503145 0.00000000 0.00000000
## omega_V 0.0000000 0.01594701 0.00000000
## omega_Cl 0.0000000 0.00000000 0.07323701
##
## States ($state or $stateDf):
## Compartment Number Compartment Name
## 1 1 depot
## 2 2 central
## ── μ-referencing ($muRefTable): ──
## theta eta level
## 1 ka_pop omega_ka id
## 2 V_pop omega_V id
## 3 Cl_pop omega_Cl id
##
## ── Model (Normalized Syntax): ──
## function() {
## description <- "The administration is extravascular with a first order absorption (rate constant ka).\nThe PK model has one compartment (volume V) and a linear elimination (clearance Cl).\nThis has been modified so that it will run without the model library"
## dfObs <- 120
## dfSub <- 12
## thetaMat <- lotri({
## ka_pop ~ 0.09785
## V_pop ~ c(0.00082606, 0.00041937)
## Cl_pop ~ c(-4.2833e-05, -6.7957e-06, 1.1318e-05)
## omega_ka ~ c(omega_ka = 0.022259)
## omega_V ~ c(omega_ka = -7.6443e-05, omega_V = 0.0014578)
## omega_Cl ~ c(omega_ka = 3.062e-06, omega_V = -1.2912e-05,
## omega_Cl = 0.0039578)
## a ~ c(omega_ka = -0.0001227, omega_V = -6.5914e-05, omega_Cl = -0.00041194,
## a = 0.015333)
## b ~ c(omega_ka = -1.3886e-05, omega_V = -3.1105e-05,
## omega_Cl = 5.2805e-05, a = -0.0026458, b = 0.00056232)
## })
## validation <- c("ipred relative difference compared to Monolix ipred: 0.04%; 95% percentile: (0%,0.52%); rtol=0.00038",
## "ipred absolute difference compared to Monolix ipred: 95% percentile: (0.000362, 0.00848); atol=0.00254",
## "pred relative difference compared to Monolix pred: 0%; 95% percentile: (0%,0%); rtol=6.6e-07",
## "pred absolute difference compared to Monolix pred: 95% percentile: (1.6e-07, 1.27e-05); atol=3.66e-06",
## "iwres relative difference compared to Monolix iwres: 0%; 95% percentile: (0.06%,32.22%); rtol=0.0153",
## "iwres absolute difference compared to Monolix pred: 95% percentile: (0.000403, 0.0138); atol=0.00305")
## ini({
## ka_pop <- 0.426994483535611
## V_pop <- -0.786351566327091
## Cl_pop <- -3.21457597916301
## a <- c(0, 0.433279557549051)
## b <- c(0, 0.0542595276206251)
## omega_ka ~ 0.450314511978718
## omega_V ~ 0.0159470121255372
## omega_Cl ~ 0.0732370098834837
## })
## model({
## cmt(depot)
## cmt(central)
## ka <- exp(ka_pop + omega_ka)
## V <- exp(V_pop + omega_V)
## Cl <- exp(Cl_pop + omega_Cl)
## d/dt(depot) <- -ka * depot
## d/dt(central) <- +ka * depot - Cl/V * central
## Cc <- central/V
## CONC <- Cc
## CONC ~ add(a) + prop(b) + combined1()
## })
## }# Look at the validation
plot(rx)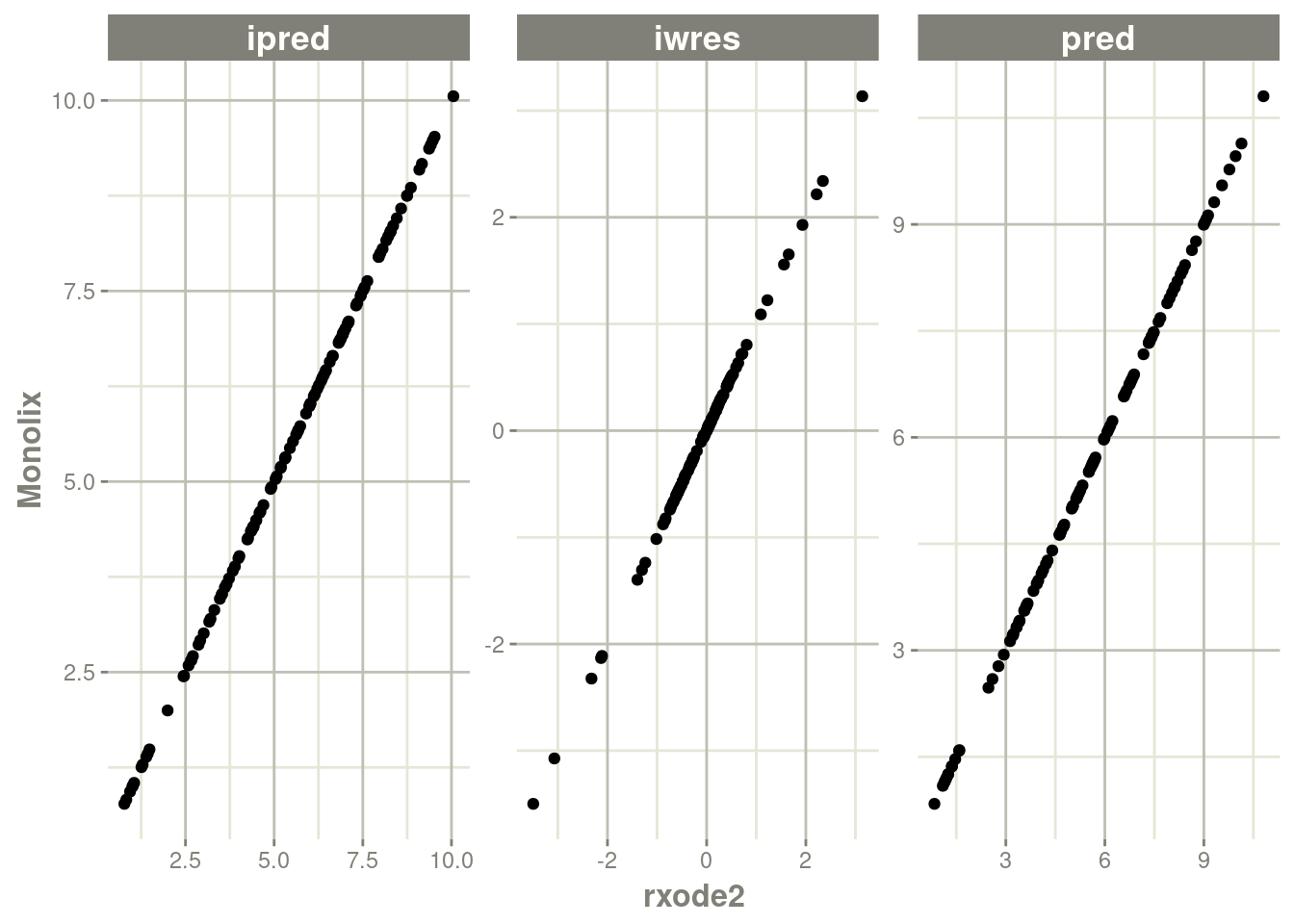
# If you want this to be converted to a nlmixr2 fit you can also
# convert it to a nlmixr2 model using babelmixr2
library(babelmixr2)
fit <- as.nlmixr2(rx)## → loading into symengine environment...## → pruning branches (`if`/`else`) of full model...## ✔ done## → finding duplicate expressions in EBE model...## [====|====|====|====|====|====|====|====|====|====] 0:00:00## → optimizing duplicate expressions in EBE model...## [====|====|====|====|====|====|====|====|====|====] 0:00:00## → compiling EBE model...## using C compiler: ‘gcc (Ubuntu 11.4.0-1ubuntu1~22.04) 11.4.0’## ✔ done## → Calculating residuals/tables## ✔ done## → compress origData in nlmixr2 object, save 7168## ℹ monolix parameter history integrated into fit objectfit## ── nlmixr² monolix2rx reading Monolix ver 5.1.1 ──
##
## OBJF AIC BIC Log-likelihood Condition#(Cov)
## monolix 118.9368 355.482 377.7819 -169.741 21.26161
## Condition#(Cor)
## monolix 1.383153
##
## ── Time (sec fit$time): ──
##
## setup optimize covariance table compress as.nlmixr2
## elapsed 0.00165 3e-06 5e-06 0.046 0.006 2.082
##
## ── Population Parameters (fit$parFixed or fit$parFixedDf): ──
##
## Est. SE %RSE Back-transformed(95%CI) BSV(CV%) Shrink(SD)%
## ka_pop 0.427 0.204 47.8 1.53 (1.03, 2.29) 75.4 1.05%
## V_pop -0.786 0.045 5.72 0.456 (0.417, 0.497) 12.7 13.3%
## Cl_pop -3.21 0.0837 2.61 0.0402 (0.0341, 0.0473) 27.6 2.65%
## a 0.433 0.433
## b 0.0543 0.0543
##
## Covariance Type (fit$covMethod): monolix2rx
## No correlations in between subject variability (BSV) matrix
## Full BSV covariance (fit$omega) or correlation (fit$omegaR; diagonals=SDs)
## Distribution stats (mean/skewness/kurtosis/p-value) available in fit$shrink
## Censoring (fit$censInformation): No censoring
## Minimization message (fit$message):
## IPRED relative difference compared to Monolix IPRED: 0.04%; 95% percentile: (0%,0.52%); rtol=0.000379
## PRED relative difference compared to Monolix PRED: 0%; 95% percentile: (0%,0%); rtol=4.94e-07
## IPRED absolute difference compared to Monolix IPRED: atol=0.00253; 95% percentile: (0.000364, 0.00848)
## PRED absolute difference compared to Monolix PRED: atol=4.94e-07; 95% percentile: (1.13e-08, 0.000308)
##
## ── Fit Data (object fit is a modified tibble): ──
## # A tibble: 120 × 20
## ID TIME DV PRED RES IPRED IRES IWRES omega_ka omega_V
## <fct> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 1 0.25 2.84 2.78 0.0636 3.73 -0.887 -1.40 0.132 -0.183
## 2 1 0.57 6.57 5.00 1.57 6.57 0.00239 0.00303 0.132 -0.183
## 3 1 1.12 10.5 6.80 3.70 8.75 1.75 1.93 0.132 -0.183
## # ℹ 117 more rows
## # ℹ 10 more variables: omega_Cl <dbl>, CONC <dbl>, depot <dbl>, central <dbl>,
## # ka <dbl>, V <dbl>, Cl <dbl>, Cc <dbl>, tad <dbl>, dosenum <dbl># This means you can do anything you could in `nlmixr2` with this
# model, even add something like conditional weighted residuals:
fit <- addCwres(fit)## → loading into symengine environment...## → pruning branches (`if`/`else`) of full model...## ✔ done## → calculate jacobian## [====|====|====|====|====|====|====|====|====|====] 0:00:00## → calculate sensitivities## [====|====|====|====|====|====|====|====|====|====] 0:00:00## → calculate ∂(f)/∂(η)## [====|====|====|====|====|====|====|====|====|====] 0:00:00## → calculate ∂(R²)/∂(η)## [====|====|====|====|====|====|====|====|====|====] 0:00:00## → finding duplicate expressions in inner model...## [====|====|====|====|====|====|====|====|====|====] 0:00:00## → optimizing duplicate expressions in inner model...## [====|====|====|====|====|====|====|====|====|====] 0:00:00## → finding duplicate expressions in EBE model...## [====|====|====|====|====|====|====|====|====|====] 0:00:00## → optimizing duplicate expressions in EBE model...## [====|====|====|====|====|====|====|====|====|====] 0:00:00## → compiling inner model...## using C compiler: ‘gcc (Ubuntu 11.4.0-1ubuntu1~22.04) 11.4.0’## ✔ done## → finding duplicate expressions in FD model...## [====|====|====|====|====|====|====|====|====|====] 0:00:00## → optimizing duplicate expressions in FD model...## [====|====|====|====|====|====|====|====|====|====] 0:00:00## → compiling EBE model...## ✔ done## → compiling events FD model...## using C compiler: ‘gcc (Ubuntu 11.4.0-1ubuntu1~22.04) 11.4.0’## ✔ done## → Calculating residuals/tables## ✔ doneprint(fit)## ── nlmixr² monolix2rx reading Monolix ver 5.1.1 ──
##
## OBJF AIC BIC Log-likelihood Condition#(Cov)
## FOCEi 120.0406 356.5859 378.8858 -170.2929 21.26161
## monolix 118.9368 355.4820 377.7819 -169.7410 21.26161
## Condition#(Cor)
## FOCEi 1.383153
## monolix 1.383153
##
## ── Time (sec $time): ──
##
## setup optimize covariance table compress as.nlmixr2
## elapsed 0.00165 3e-06 5e-06 0.046 0.006 2.082
##
## ── Population Parameters ($parFixed or $parFixedDf): ──
##
## Est. SE %RSE Back-transformed(95%CI) BSV(CV%) Shrink(SD)%
## ka_pop 0.427 0.204 47.8 1.53 (1.03, 2.29) 75.4 1.05%
## V_pop -0.786 0.045 5.72 0.456 (0.417, 0.497) 12.7 13.3%
## Cl_pop -3.21 0.0837 2.61 0.0402 (0.0341, 0.0473) 27.6 2.65%
## a 0.433 0.433
## b 0.0543 0.0543
##
## Covariance Type ($covMethod): monolix2rx
## No correlations in between subject variability (BSV) matrix
## Full BSV covariance ($omega) or correlation ($omegaR; diagonals=SDs)
## Distribution stats (mean/skewness/kurtosis/p-value) available in $shrink
## Censoring ($censInformation): No censoring
## Minimization message ($message):
## IPRED relative difference compared to Monolix IPRED: 0.04%; 95% percentile: (0%,0.52%); rtol=0.000379
## PRED relative difference compared to Monolix PRED: 0%; 95% percentile: (0%,0%); rtol=4.94e-07
## IPRED absolute difference compared to Monolix IPRED: atol=0.00253; 95% percentile: (0.000364, 0.00848)
## PRED absolute difference compared to Monolix PRED: atol=4.94e-07; 95% percentile: (1.13e-08, 0.000308)
##
## ── Fit Data (object is a modified tibble): ──
## # A tibble: 120 × 24
## ID TIME DV PRED RES IPRED IRES IWRES omega_ka omega_V
## <fct> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 1 0.25 2.84 2.78 0.0636 3.73 -0.887 -1.40 0.132 -0.183
## 2 1 0.57 6.57 5.00 1.57 6.57 0.00239 0.00303 0.132 -0.183
## 3 1 1.12 10.5 6.80 3.70 8.75 1.75 1.93 0.132 -0.183
## # ℹ 117 more rows
## # ℹ 14 more variables: omega_Cl <dbl>, CONC <dbl>, depot <dbl>, central <dbl>,
## # ka <dbl>, V <dbl>, Cl <dbl>, Cc <dbl>, tad <dbl>, dosenum <dbl>,
## # WRES <dbl>, CPRED <dbl>, CRES <dbl>, CWRES <dbl>- Posted on:
- October 21, 2024
- Length:
- 11 minute read, 2239 words
- Categories:
- rxode2 monolix2rx monolix babelmixr2
- See Also: